Daily Kos bags of words from the time of the 2004 Presidential election
This is a bit of a rambly blog entry today. My original motivation was to just explore moving data around from R into the H2O machine learning software. While successful on this, and more on it below, I got a bit interested in my example data in its own right. The best initial exploratory analysis of that data turns out on my opinion not to be one of the approaches implemented in H2O, so by the end of the post I’m back in R illustrating the use of topic modelling for analysing blog posts. As a teaser, this is where we’re going to end up:
That graphic represents eight latent “topics” identified in 3,430 posts on the liberal (as per US terminology) Daily Kos site from the time of the 2004 US Presidential Election. The data are available at the Machine Learning Repository of the Univerity of California’s Center for Machine Learning and Intelligent Systems. The metadata are low on context (or perhaps I was just looking in the wrong spot) - the origin is just described as “KOS blog entries … orig source: dailykos.com”, and I’m presuming from their content that they are from 2004. More on what it all means down below.
Sparse matrices
I chose this data because it’s a nice example of a medium sized sparse matrix. The data has the number of occurences of 6,906 words in 3,430 documents so the obvious conceptual format is a matrix with 3,430 rows and 6,906 columns, where most cells are empty (meaning zero occurences of that word in that document) and the filled cells are counts. Of course, we try to avoid moving data around like that. Instead of that super-wide matrix, the data is distributed as a text file with three columns that looks like this:
1 61 2
1 76 1
1 89 1
1 211 1
1 296 1
1 335 1
1 404 1
1 441 1
1 454 2In this case, it means that the first document has two counts of word 61; one count of word 76; one count of word 89; and so on. A separate file tells us that word 61 is “action”, word 76 is “added” and so on. In fact, the two files for distributing these data are just the two tables that you’d represent them with in a relational database. This is the optimally efficient way of storing data of this sort.
Now, I’ve been exploring the machine learning software H2O which provides fast and efficient estimation of models such as :
- deep learning neural networks
- random forests
- gradient boosting machines
- generalized linear models with elastic net regularisation
- k-means clustering
All these tools need data to be represented in a wide format, rather than the efficient long format you’d have in a relational database. In this case, that would mean a row for each document (or observation) and a column for each word, which would be used as “features” in machine learning terminology. That is, it needs the sort of operation you’d use PIVOT for in SQL Server, or tidyr::spread in R (or, before the coming of the tidyverse, reshape or reshape2::cast).
H2O is expected to work with another tool for data persistence eg a Hadoop cluster. One thing I’m still getting my head around is the several variants on making data actually available to H2O. I’m particularly interested in the case of medium-sized data that can be handled on a single machine in efficient sparse format, when a Hadoop cluster isn’t available. So the Daily Kos data was my small familiarisation example in this space.
A common workflow I expect with this sort of data is a bunch of pre-processing that will need a general data munging tool like R or Python before upload into H2O. For example, with text data, I might want to do one or more of remove common “stopwords” (like ‘the’, ‘and’, etc); reduce words to their “stems” (so “abandons” and “abandon” are treated as the same); match words to sentiment; etc. This is the sort of thing done with NLTK in Python or tm, SnowballC and the brilliant new tidytext in R.
tidytext by Julia Silge and David Robinson makes it possible to analyse textual data using the general data manipulation grammar familiar to users of Hadley Wickham’s tidyverse. One verb tidytext offers is cast_sparse, which converts data in long tidy format into a wide but sparse Matrix format (using Doug Bates and Martin Maechler’s Matrix package, which is distributed with base R) that is suitable for a range of statistical and machine learning models. So I was pleased to note that the latest stable version of H2O (not yet on CRAN at the time of writing) and its accompanying R interface expands the h2o::as.H2O function to support the sparse Matrix format.
Importing and modifying the data
So here’s how I set about exploring a workflow based on the following key tools
- Pre-processing in R
- Import data with
data.table::fread- the fastest way to get data in. Whilereadror evenutilsfunctions would also do the job well in this modest-sized case, I want to scale up in the future andfreadis the best there is for getting large, regular text data into R. - Modify with tools from the
tidyverseandtidytext, calling in specialist packages for stemming etc as necessary - Cast to sparse
Matrixformat and export to H2O withh2o::as.h2o
- Import data with
- Analyse in H2O
- Export the summary results back to R for presentation
I had my doubts about whether h2o::as.h2o is right for the job, because I understand it doesn’t make the most of H2O’s own fast data importing abilities. But it is definitely a candidate for medium sized data.
Here’s how I got started. This first chunk just loads the functionality used in the rest of the post and downloads the main table, if it hasn’t been downloaded before (I developed this script in little chunks of time between doing other stuff, and didn’t want to download the file every time I ran it or have to bother with selectively running bits of the script). Lots of R packages needed for today’s exploration:
library(ggplot2)
library(scales)
library(R.utils) # for gunzip
library(tidyverse)
library(tidytext)
library(Matrix) # for sparse matrices
library(data.table) # for fread, fast version of read.table
library(SnowballC) # for wordStem
library(foreach)
library(doParallel)
library(testthat) # used for various checks and tests along the way
library(topicmodels)
library(slam) # for other types of sparse matrices, used with topicmodels
library(wordcloud)
library(grid) # for annotating graphics
# latest version of h2o needed at time of writing (December 2016) for sparse matrix support
# install.packages("h2o", type="source", repos=(c("http://h2o-release.s3.amazonaws.com/h2o/rel-tutte/1/R")))
library(h2o)
# https://archive.ics.uci.edu/ml/datasets/Bag+of+Words
# download the smallest example bag of words for first trial
if(!"docword.kos.txt" %in% list.files()){
download.file("https://archive.ics.uci.edu/ml/machine-learning-databases/bag-of-words/docword.kos.txt.gz",
destfile = "docword.kos.txt.gz", mode = "wb")
gunzip("docword.kos.txt.gz")
}Importing the data is super fast with data.table::fread. Combining it with the vocabulary look up table, removing stop words and performing stemming is made super-easy by tidytext in combination with text mining specialist tools, in this case SnowballC::wordStem:
kos <- fread("docword.kos.txt", skip = 3, sep = " ")
names(kos) <- c("doc", "word", "count")
expect_equal(sum(kos$count), 467714)
kos_vocab <- read.table("https://archive.ics.uci.edu/ml/machine-learning-databases/bag-of-words/vocab.kos.txt")$V1
kos2 <- kos %>%
# attach the names of words back to the words
mutate(word = factor(word, labels = kos_vocab)) %>%
# stopwords are meant to be already removed but no harm in checking (note - makes no difference):
anti_join(stop_words, by = "word") %>%
# reduce words to their stems (otherwise abandon, abandoning, abandons all treated as different, etc),
mutate(word = wordStem(word)) %>%
mutate(word = gsub("^iraq$", "iraqi", word)) %>%
# ... and aggregate back up by the new stemmed words to remove duplicates
group_by(doc, word) %>%
summarise(count = sum(count)) %>%
# correct some names that went wrong in the stemming:
mutate(word = gsub("^chenei$", "cheney", word),
word = gsub("^kerri$", "kerry", word),
word = gsub("^georg$", "george", word),
word = gsub("^john$", "John", word))
# how many words per post?
wordcount <- kos2 %>%
group_by(doc) %>%
summarise(words = sum(count))
par(font.main = 1)
hist(wordcount$words, col = "grey", fg = "grey80", border = "grey95",
main = "Words per post in the KOS data set", xlab = "Number of words")Because the post lengths varied in the predictable way (histogram above), I decided to convert all the counts to proportions of words in each original document. The next chunk does this, and then casts the data out of long format into wide, analysis-ready format. I create both sparse and dense versions of the wide matrix to check that things are working the way I want (this is one of the reasons for checking things out on a fairly small dataset). In the chunk below, note that I had a bit of a fiddle (and then built in a test to check I got it right) to get the columns in the same order with cast_sparse as when I used spread for the dense version.
# OK, quite a variety of words per post.
# Convert counts to *relative* count (ie proportion of words in each doc)
# need to do this as otherwise the main dimension in the data is just the
# length of each blog post
kos3 <- kos2 %>%
group_by(doc) %>%
mutate(count = count / sum(count))
# Sparse version
kos_sparse <- kos3 %>%
# arrange it so the colnames after cast_sparse are in alphabetical order:
arrange(word) %>%
cast_sparse(doc, word, count)
# Dense version
kos_df <- kos3 %>%
spread(word, count, fill = 0)
kos_dense <- as.matrix(kos_df[ , -1])
expect_equal(colnames(kos_dense), colnames(kos_sparse))
dim(kos_sparse) # would be 3430 documents and 6906 words except that there's been more processing, so only 4566 words
object.size(kos) # about 4MB
object.size(kos_sparse) # about the same. Slightly less efficient
object.size(kos_dense) # 120+ MBk-means cluster analysis in R
As this exercise started as essentially just a test of getting data into H2O, I needed an H2O algorithm to test on the data. With just bags of words and no other metadata (such as how popular each post was, or who wrote it, or anything else of that sort) there’s no obvious response variable to test predictive analytics methods on, so I chose k-means clustering as my test algorithm instead. There’s reasons why this method isn’t particularly useful for this sort of high-dimensional (ie lots of columns) data, but they’re not enough to stop it being a test of sorts.
One of the challenges of cluster analysis is to know how many groups (k) to divide the data into. So give R and H2O a decent run, I applied the k-means clustering to the data (using the dense version of the matrix) with each value of k from 1 to 25.
Close readers - if there are any - may note that I haven’t first scaled all the columns in the matrix before applying the k-means algorithm, even though this is often recommended to avoid columns with high variance dominating the clustering. This is a deliberate choice from me, applicable to this particular dataset. As all the columns are on the same scale (“this word’s frequency as a proportion of total words in the document for this row”) I thought it made sense to let the high variance columns sway the results, rather than (in effect) let low variance columns for rarely-used words get equal votes.
#========k-means cluster analysis in R===========
# set up parallel cluster for the R version of analysis
# (sorry for there being two uses of the word "cluster"...)
cluster <- makeCluster(7) # only any good if you have at least 7 processors :)
registerDoParallel(cluster)
clusterExport(cluster, "kos_dense")
# How to work out how many clusters to find in the data?
# great answer at http://stackoverflow.com/questions/15376075/cluster-analysis-in-r-determine-the-optimal-number-of-clusters
# But I only use the simplest method - visual inspection of a scree plot of the total within squares
# takes some time, even with the parallel processing:
max_groups <- 25
wss <- foreach (i = 1:max_groups) %dopar% {
kmeans(kos_dense, centers = i)$tot.withinss
}
par(font.main = 1)
plot(1:max_groups, wss, type="b", xlab="Number of groups",
ylab="Within groups sum of squares", bty = "l")
grid()
stopCluster(cluster)This graphic shows the total within-groups sum of squares for each value of k. The within-groups sum of squares is an indicator of how homogenous the points within each group are. Inevitably it goes down as the number of groups dividing the data increases; we draw plots like this one to see if there is a visually-obvious “levelling-off” point to indicate when we are getting diminishing marginal returns from adding more groups.
I won’t worry too much about interpreting this as I think the method is of dubious value anyway. But my gut feel is that either there is 1, 2, or about 20 groups in this data.
Getting the data into H2O
For the H2O comparison, first step is to initiate an H2O cluster and get the data into it. With the newly enhanced as.h2o function working with sparse Matrix objects this is easy and takes only 3 seconds (compared to 22 seconds for uploading the dense version of the matrix, code not shown).
There’s a funny quirk to watch out for; the column names don’t get imported, and only 100 column names are applied.
I suspect the colnames issue after importing a sparse Matrix to H2O is a bug. but will make sure before I report it as an issue.
Having imported the data, I do a basic check. k-means with a single group should return just the mean of each column as the p-dimensional centroid of a single big hyper-sphere containing all the data. I want to check that the various versions of the data in R and H2O all return the same values for this.
#==============cluster analysis in h2o======================
h2o.init(nthreads = -1, max_mem_size = "10G")
system.time({ kos_h1 <- as.h2o(kos_sparse)}) # 3 seconds
dim(kos_h1) # correct
colnames(kos_h1) # for some reason, only 100 columnames, c1:100
colnames(kos_h1) <- colnames(kos_sparse)
#--------------------checking we have the same data in all its different versions--------------------
# Compare the simplest single cluster case - should just be the mean of each column
simple_r <- kmeans(kos_dense, centers = 1)
simple_h <- h2o.kmeans(kos_h1, x = colnames(kos_h1), k = 1, standardize = FALSE)
simple_m1 <- apply(kos_dense, 2, mean)
simple_m2 <- t(apply(kos_h1, 2, mean))
simple_m3 <- apply(kos_sparse, 2, mean)
comparison <- data.frame(
R = simple_r$centers[1 ,],
manual1 = simple_m1,
H2O = t(h2o.centers(simple_h))[,1],
manual2 = as.vector(simple_m2[,1]),
manual3 = simple_m3
)
par(font.main = 1)
pairs(comparison, main = "Different methods of estimating the center of the data")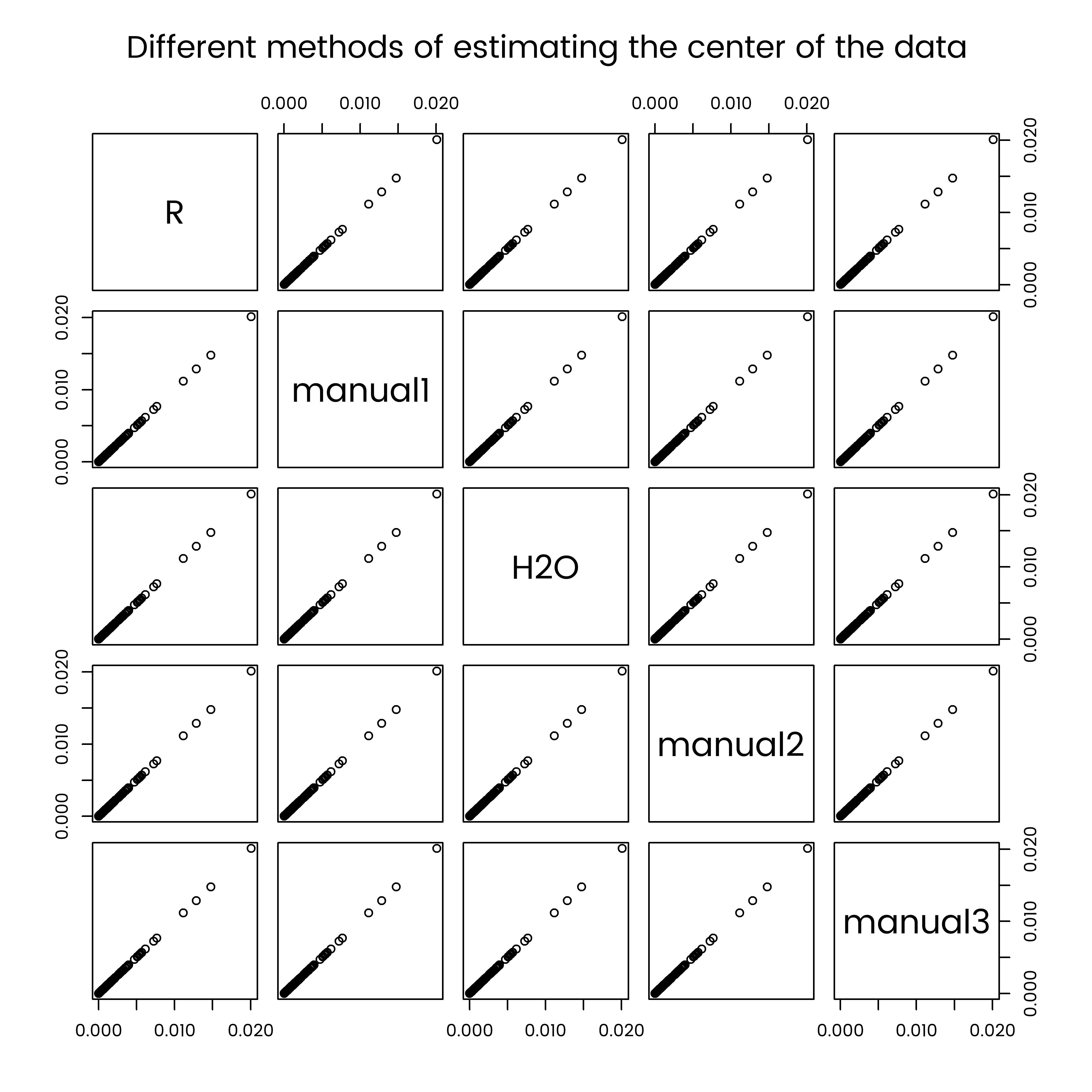
And the result is fine. The data import has worked ok. Note that this wasn’t the case when I first did it - after a while I tracked it down to the non-alphabetical ordering of columns created by cast_sparse, which I have now forced to be alphabetical in the code earlier in this post.
k-means cluster analysis in H2O
To complete my experiment, I ran the k-means clustering algorithm on the data in H2O in a similar way to how I’d already done it in base R. One advantage of H2O in this context is it lets you use cross-validation for assessing the within-group sum of squares, rather than getting it from the data that was used to set the centers. This takes longer but is a better way of doing it. In fact, I tried it with and without cross validation and got similar results - the cross-validation total within groups sum of squares is always a little higher than that from the training data, but not enough to disrupt patterns.
max_groups <- 25
wss_h2o <- numeric(max_groups)
for(i in 1:max_groups){
kos_km1 <- h2o.kmeans(kos_h1, x= colnames(kos_h1), estimate_k = FALSE, k = i, standardize = FALSE,
nfolds = 5, max_iterations = 25)
wss_h2o[i] <- h2o.tot_withinss(kos_km1, xval = TRUE) # xval=TRUE means cross validation used
}
par(font.main = 1)
plot(1:max_groups, wss_h2o, type="b", xlab="Number of Clusters",
ylab="Within groups sum of squares", bty = "l")
grid()Note that the scree plot from H2O is much less regular than that from base R. This was the case whether it was created with cross-validation values of total within group sum of squares, or those from the training data. Basically, the H2O k-means algorithm appears to be a little less steady than that in R. This would worry me except for my concerns with the appropriateness of the method at all to this sort of data. Because of the “curse of dimensionality”, nearly all points in this 4,000 dimensional space are pretty much equally far from each-other, and it’s not surprising that a technique that aims to efficiently group them into clusters finds itself in unstable territory.
So I’m not going to spend more space on presenting the final model, as I just don’t think the method is a good one in this case. The exercise has served its purpose of showing that the new as.h2o upload of sparse Matrix format data into H2O works. Unfortunately, it didn’t scale up to larger data. The New York Times bags of words in the same repository as the Kos Blog data has about 300,000 documents and a vocabulary of 100,000 words, and although R, tidytext and Matrix handle it fine I couldn’t upload to H2O with as.h2o. So this remains an open problem. If you know how to fix it, please answer my question on Stack Overflow! The solution could be either a way of pivoting/spreading long form data in H2O itself, or a better way of handling the upload of pre-pivoted wide sparse data. Or perhaps I just need more computing power.
Topic modelling in R
I would have stopped there but I thought it was a waste of a good dataset to use only an admittedly inferior method to analyse it. So I reached into the toolbox and grabbed the topic modelling approach, which is a more appropriate tool in this case. Topic modelling is a Bayesian approach that postulates a number of unseen latent “topics”. Each topic has its own probability distribution to generate a bag of words. The probability of each estimated topic generating each observed bag of words conditional on the best estimate of topics is considered, the process improved iteratively, and eventually one is left with a bunch of topics each of which has a probability of generating each word.
As an inferential method I think it’s over-rated, but as an exploratory approach to otherwise intractable large bunches of bags of words it’s pretty useful - sort of the equivalent exploratory analytical/graphical combined method (in my view) to the armoury of density plots, scatter plots and line charts that we use when first exploring continuous data. Oddly enough, the results are usually presented as tables of data listing the words most characteristic of each topic. I think we can do better than that… here’s the graphic from the top of the post again, for convenience:
Word clouds can be a silly gimmick, but if used well they are a good use of space to pack text in. Like any graphic, they should facilitate comparisons (in this case, between different topics).
My interpretation of these topics?
- Topics 1 and 8 might be the more tactically focused discussion of polls and punditry in the leadup to the actual election
- Topic 2 stands out clearly as the Iraq War topic
- Topic 6 clearly relates to the broader war on terror, intelligence and defence issues
- Topic 5 looks to be the general “Bush Administration” discussions
- Topic 3 seems to be focused on the Democratic primary process
- Topic 4 might be discussion of non-Presidential (Senate, House, Governor) races
- Topic 7 might be a more general discussion of the Democratic party, people, politics and parties
- The Daily Kos writers are a pretty nationally focused lot. A political blog in Australia or New Zealand might discuss elections in the USA, Britain, and probably France, Germany, Japan and maybe Indonesia as well as their own country, but the Daily Kos has the luxury of focusing on just the USA.
Overall, it’s not a bad overview of the subject-matter of 3,400 articles that the computer has read so we don’t need to.
As this is a standard and nicely curated dataset I thought there would be stacks of people who have done this before me but on a cursory search I couldn’t find it. Here are a couple of bits of related analysis:
There’s a fair bit of discussion and not much clarity on the web on how to choose the correct number of topics for a given corpus of text. I think I’ll save that for another time.
Here’s the R code that created it, no frills or playing around with arguments. Note that I had to put the data into another sparse matrix format for the excellent topicmodels R package by Bettina Gruen to work with. I use the kos2 object defined earlier in this post, which was the data after stemming and re-aggregation, but before word frequencies were converted to counts. The most common form of topic modelling uses a method called latent Dirichlet allocation which unless I’m mistaken does that conversion under the hood; but the topicmodels::LDA function I use below needs to be provided counts, not proportions.
kos_counts <- kos2 %>%
arrange(word) %>%
# take advantage of factor() as a quick way to make number:level pairings
mutate(word = factor(word))
# convert to triplet (row, column, cell) sparse matrix format:
kos_trip <- with(kos_counts, simple_triplet_matrix(i = doc, j = as.numeric(word), v = count))
dimnames(kos_trip)[[2]] <- levels(kos_counts$word)
# EDIT - on Twitter, Julia Silge pointed out these last two lines could have
# stayed in the tidytext world by adding cast_dtm() to my pipeline above. I'd
# overlooked the existence of cast_dtm(), which is indeed exactly what I was looking for.
# function for drawing word clouds of all topics from an object created with LDA
wclda <- function(lda, n = 100, palette = "Blues", ...){
p <- posterior(lda)
w1 <- as.data.frame(t(p$terms))
w2 <- w1 %>%
mutate(word = rownames(w1)) %>%
gather(topic, weight, -word)
pal <- rep(brewer.pal(9, palette), each = ceiling(n / 9))[n:1]
for(i in 1:ncol(w1)){
w3 <- w2 %>%
filter(topic == i) %>%
arrange(desc(weight))
with(w3[1:n, ],
wordcloud(word, freq = weight, random.order = FALSE,
ordered.colors = TRUE, colors = pal, ...))
title(paste("Topic", i))
}
}
system.time(lda3 <- LDA(kos_trip, k = 8)) # about 300 seconds
par(mfrow = c(2, 4), font.main = 1)
wclda(lda3)
grid.text(0.5, 0.55, label = "Latent topics identified in 3,430 dailykos.com blog posts in 2004",
gp = gpar(fontface = "italic"))
grid.text(0.98, 0.02, hjust = 1,
label = "Source: data from https://archive.ics.uci.edu/ml/datasets/Bag+of+Words\n analysis from https://ellisp.github.io",
gp = gpar(fontface = "italic", cex = 0.7, col = "grey60"))


